Where is the raw sequencing data stored publicly?
Data availability
Most peer-reviewed journals require studies dealing with bioinformatic data to include a data availability statement. This statement explains where the raw sequencing data and all important associated data sets are available. Publishing sequence data online is now mandatory and considered good practice, as it allows other scientists to repeat the analyses or use the data for their own studies.
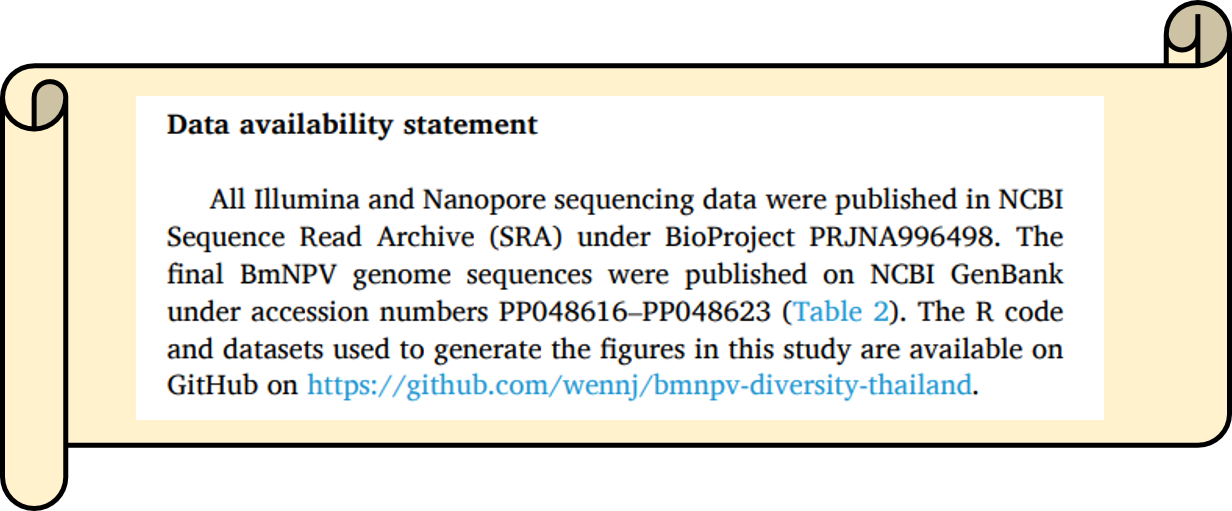
Typically, the Data Availability Statement is located at the end of the manuscript. Sometimes it is also located at the very beginning. The information can also be integrated into the Materials and Methods section.
Sequence Read Archive (SRA)
Sequence data is published as raw data. The world’s largest public database for raw sequence data from high-throughput sequencing is the Sequence Read Archive (SRA). The SRA was founded in 2007 and is now jointly operated by the NCBI (USA), the EBI/ENA (Europe), and the DDBJ (Japan).
The SRA is the world’s largest collection of sequence data.
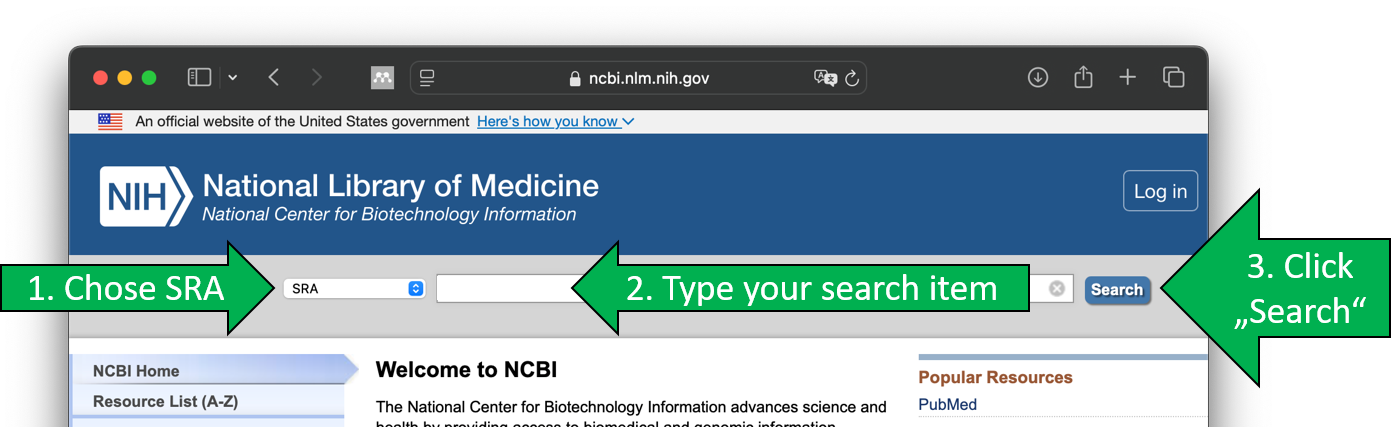
- Visit https://www.ncbi.nlm.nih.gov/sra
- Search for SRA data entry
SRR14313721
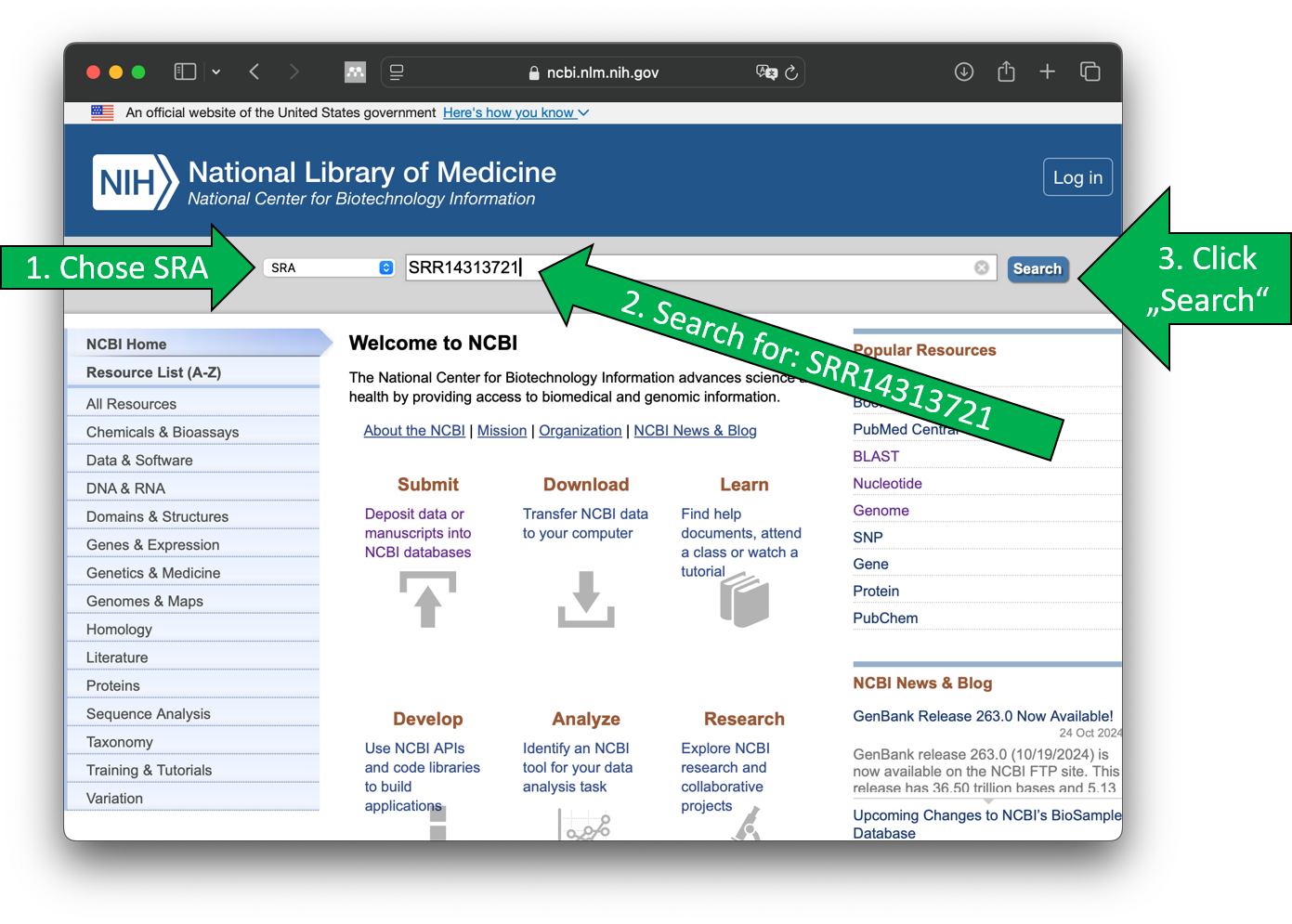 Next, you should see the SRA record entry.
Next, you should see the SRA record entry.
What information can you see?
Can you find out the meaning of…
- BioProject
- BioSample
- Run
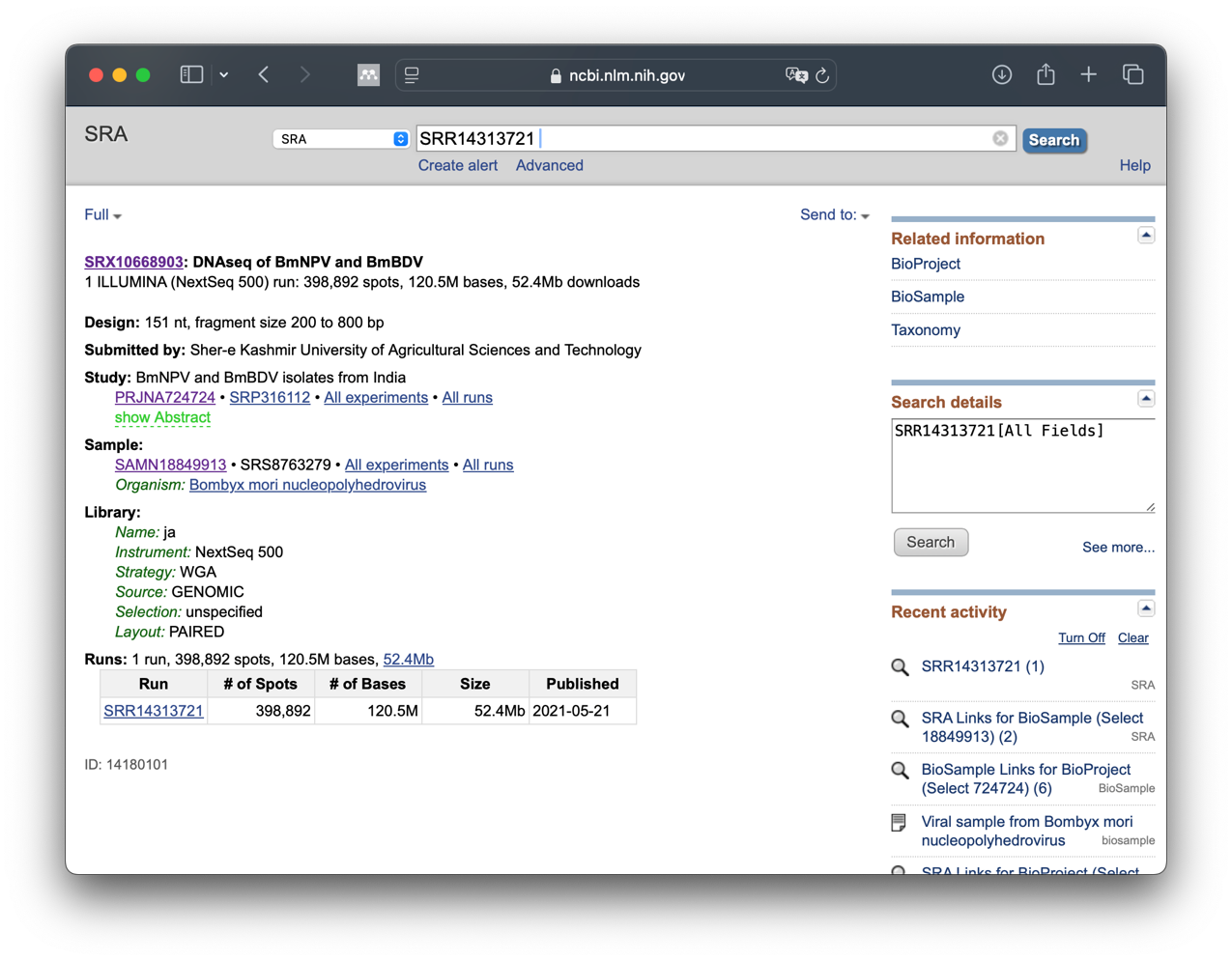
SRA entry information
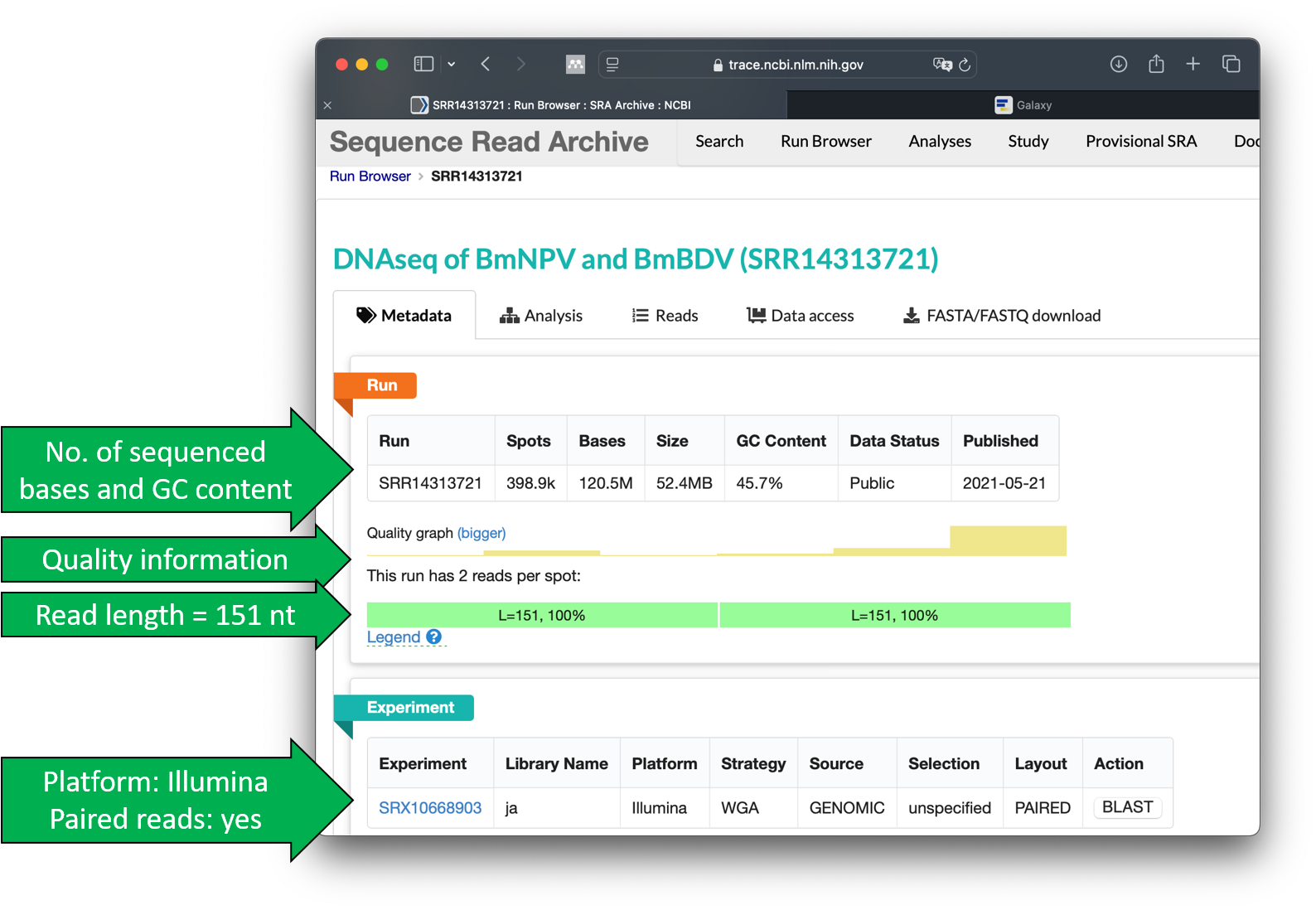
The SRA offers the possibility to retrieve a lot of information about the sequencing. We will look at this in the next step. To do this, we click directly on the run entry SRR14313721.
Click on SRR14313721.
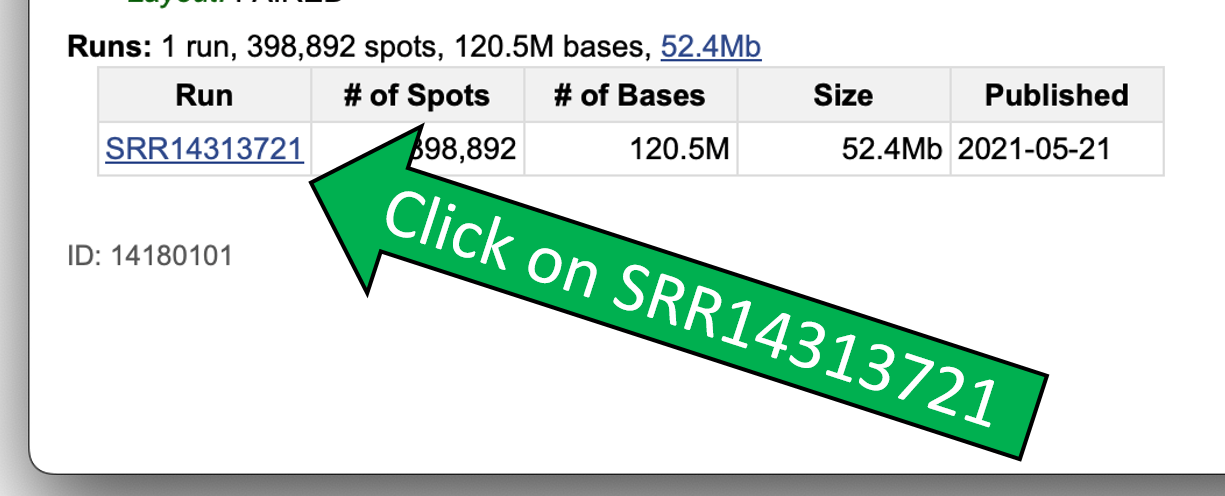
This should open the detailed view, which provides information on the number of sequenced bases, the quality of the sequencing, and the format of the sequence data.
Taxonomy Analysis Tool
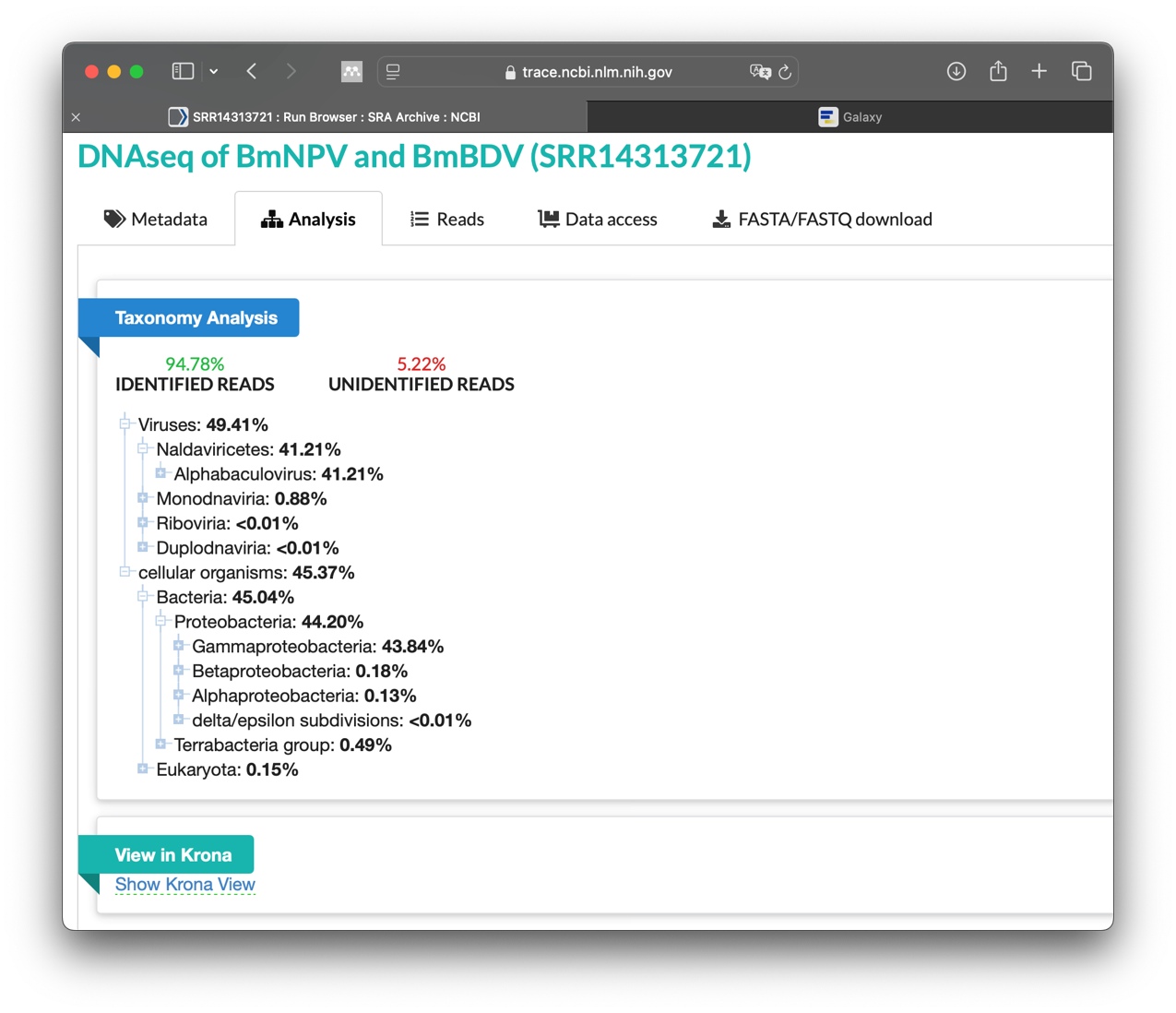
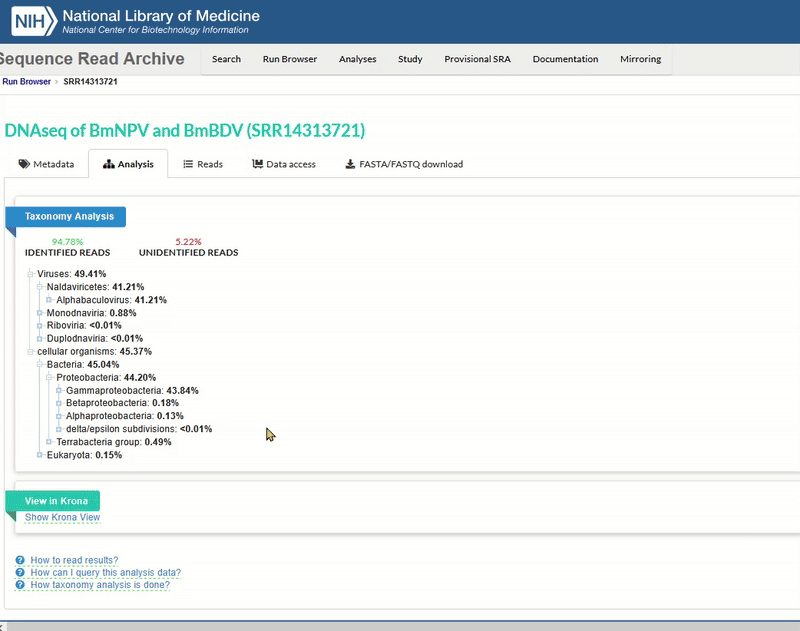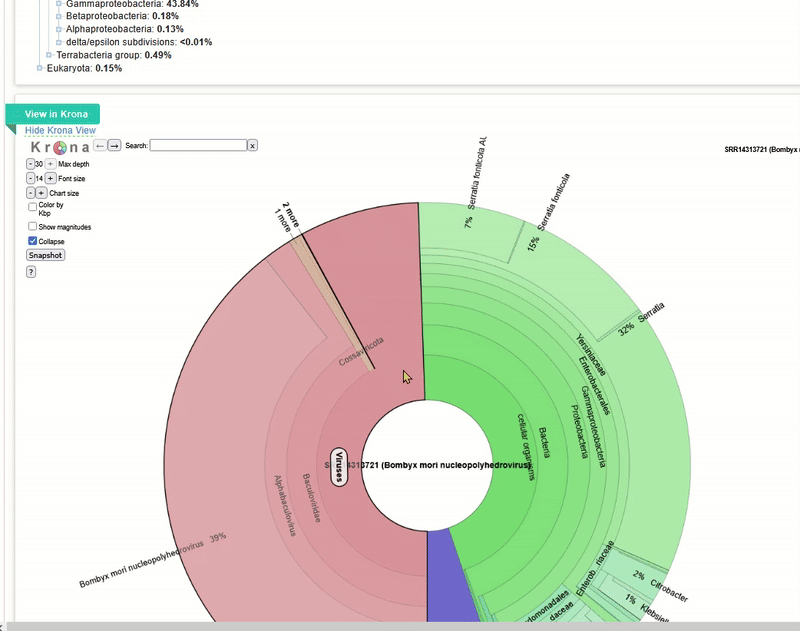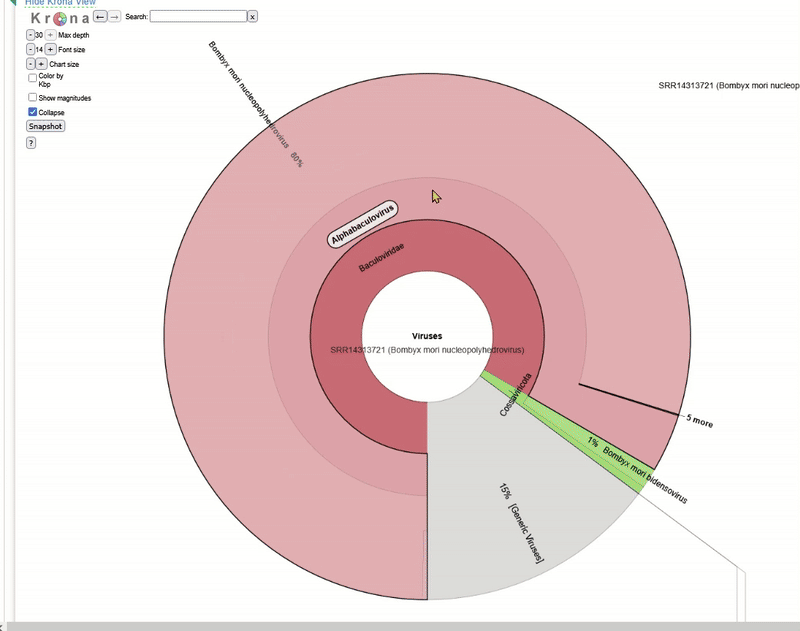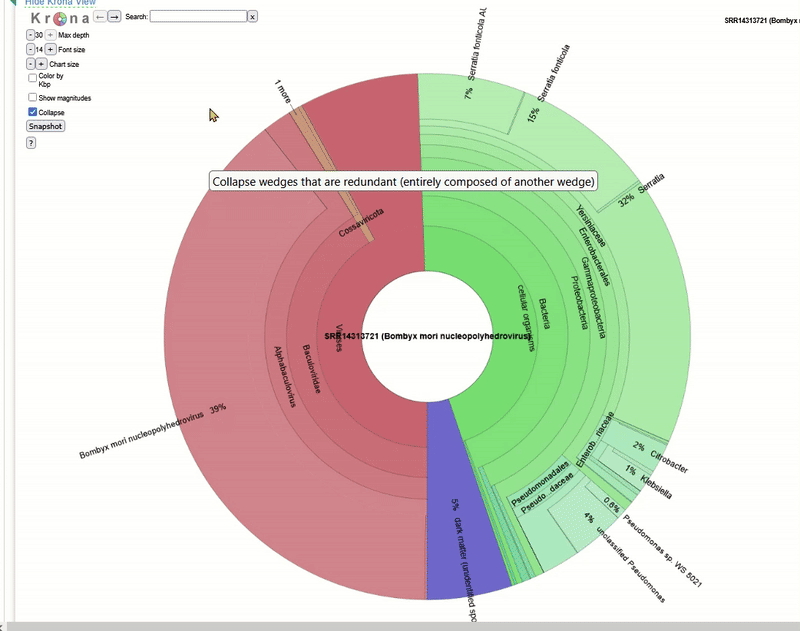
The SRA web interface also offers a simple analysis of the taxonomy. This is helpful if you want to know what is included in the sequencing. It can provide an indication of whether there may be contamination in the sequencing.
At the bottom, there is a field called ‘View in Krona’, which we click on.
 The Taxonomy Browser window will then open. This analysis is performed automatically by NCBI SRA for every dataset that is uploaded there. It is a rough metagenomic analysis based on the information stored at NCBI itself. You can see which organisms (including viruses) were found in the sequence data.
The Taxonomy Browser window will then open. This analysis is performed automatically by NCBI SRA for every dataset that is uploaded there. It is a rough metagenomic analysis based on the information stored at NCBI itself. You can see which organisms (including viruses) were found in the sequence data.
 The web interface also offers an interactive way to analyse the composition. This tool is called
The web interface also offers an interactive way to analyse the composition. This tool is called Krona, which we will launch next.

The Krona tool for interactive analysis will then open. Play around with the interactive display to understand how Krona works!
Search SRA database
It’s time to put what you’ve learned into practice. How about searching the SRA database yourself? Can you think of any organisms that might have been sequenced?
Let’s find out. Search for organisms you work with!
Which organism comes to your mind? Then search for it!
Examples:
- Ascovirus
- Homo sapiens
- Beauveria bassiana
- Silkworm
Visit https://www.ncbi.nlm.nih.gov/sra to start your search.
Also use Krona to analyse your search!
Who found something interesing?
Can you share your findings?