Principle of Illumina sequencing
Illumina sequencing (short read sequencing)
- one of the Next Generation Sequencing (NGS) technologies
- Most widely applied sequencing technique
- Generates not one but millions of sequences at the same time
- read = single sequence
- high accuracy (>99.9%)
Nucleotides with reversible terminated 3’-OH group


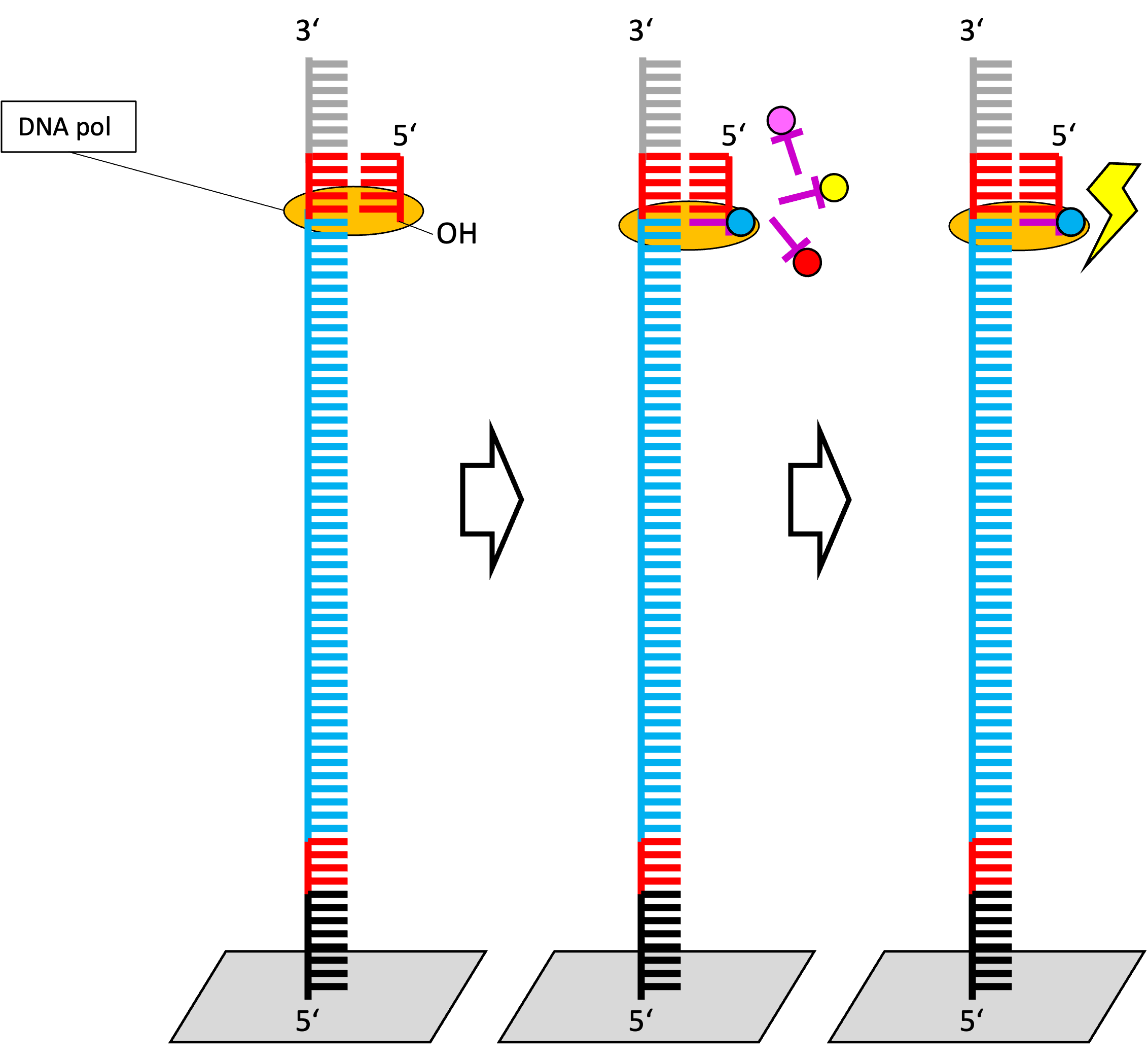
Sequencing by synthesis - step by step

Assumptions:
- Blue: Single stranded temple = sequence of interest
- The DNA strand is fixed to a surface at the 5’-OH end.
- Two terminal adapters (of known sequence) are ligated to both ends of the DNA strand.
- A primer (oligonucleotide) is bound at one side.
- Oligonucleotide as a free 3’ OH group.


- Nucleotides with reversiblee terminating groups are equally frequent in the reaction.
- DNA polymerase incorporates a single complementary nucleotide (matching wit the template).
- Fluorescent dye acts as a terminator of polymerization (=strand synthesis).
- Strand synthesis is terminated.

- The excitation of the fluorescent dye happen at a specific wavelength.
- The detection of the light emission can be used to identify the nucleotide.


- Next, the fluorescent dye is removed by enzymatic cleavage.
- The exposure of free 3’ OH group enables the elongation with the next ddNTP.
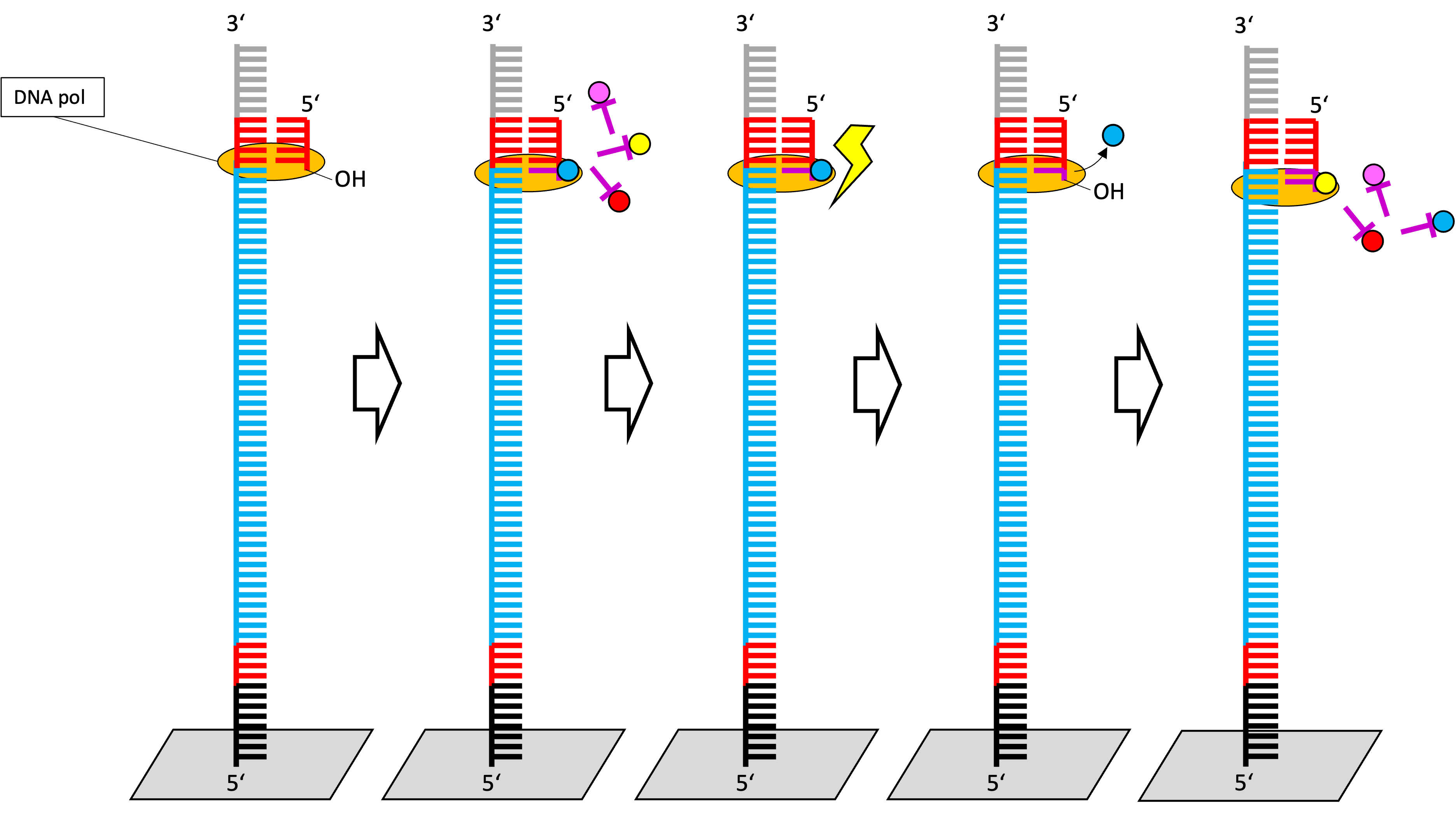

Which bases have been sequenced?


From top we see only the emitted light

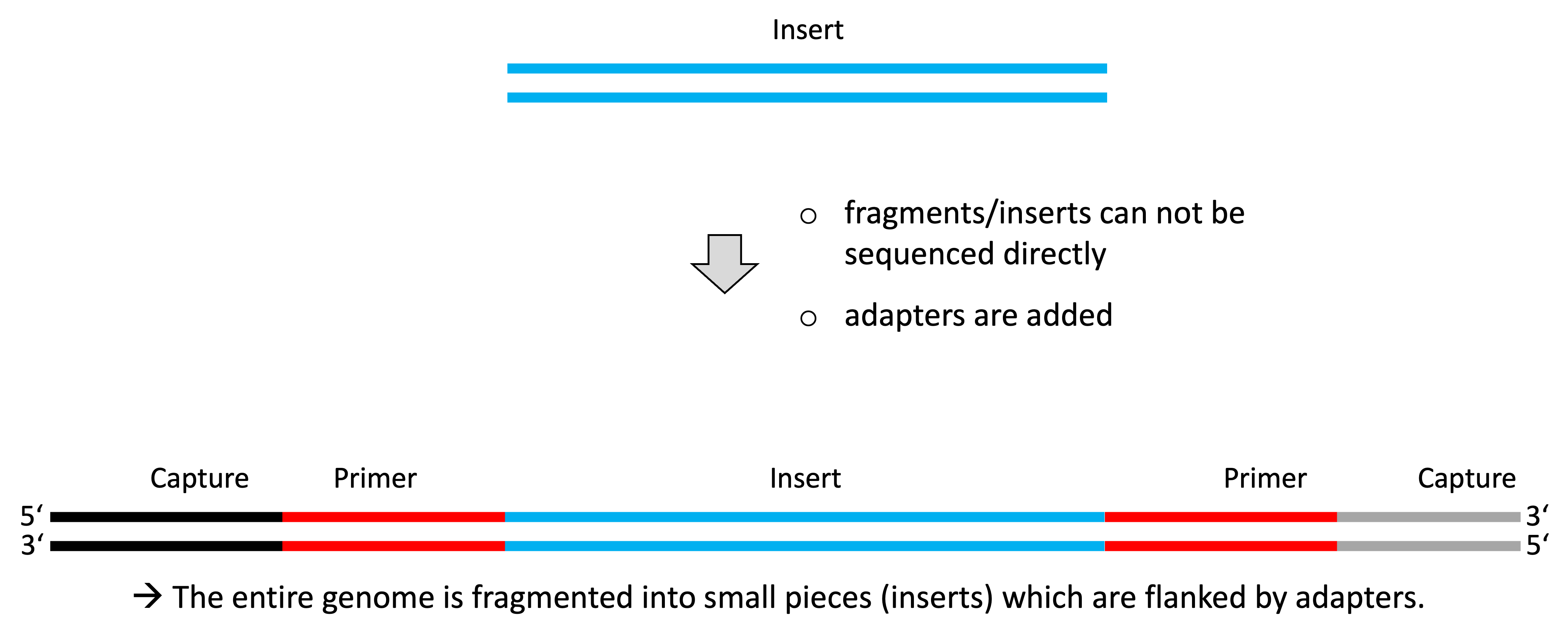
DNA fragmentation prior to sequencing

- Usually DNA is too large to be sequenced directly.
- DNA is fragmented (enzymatically/mechanically) into smaller pieces.
- Fragment sizes differ depending on the applied protocol.
- Adapters with known sequences are added to the fragments.
Sequencing adapter
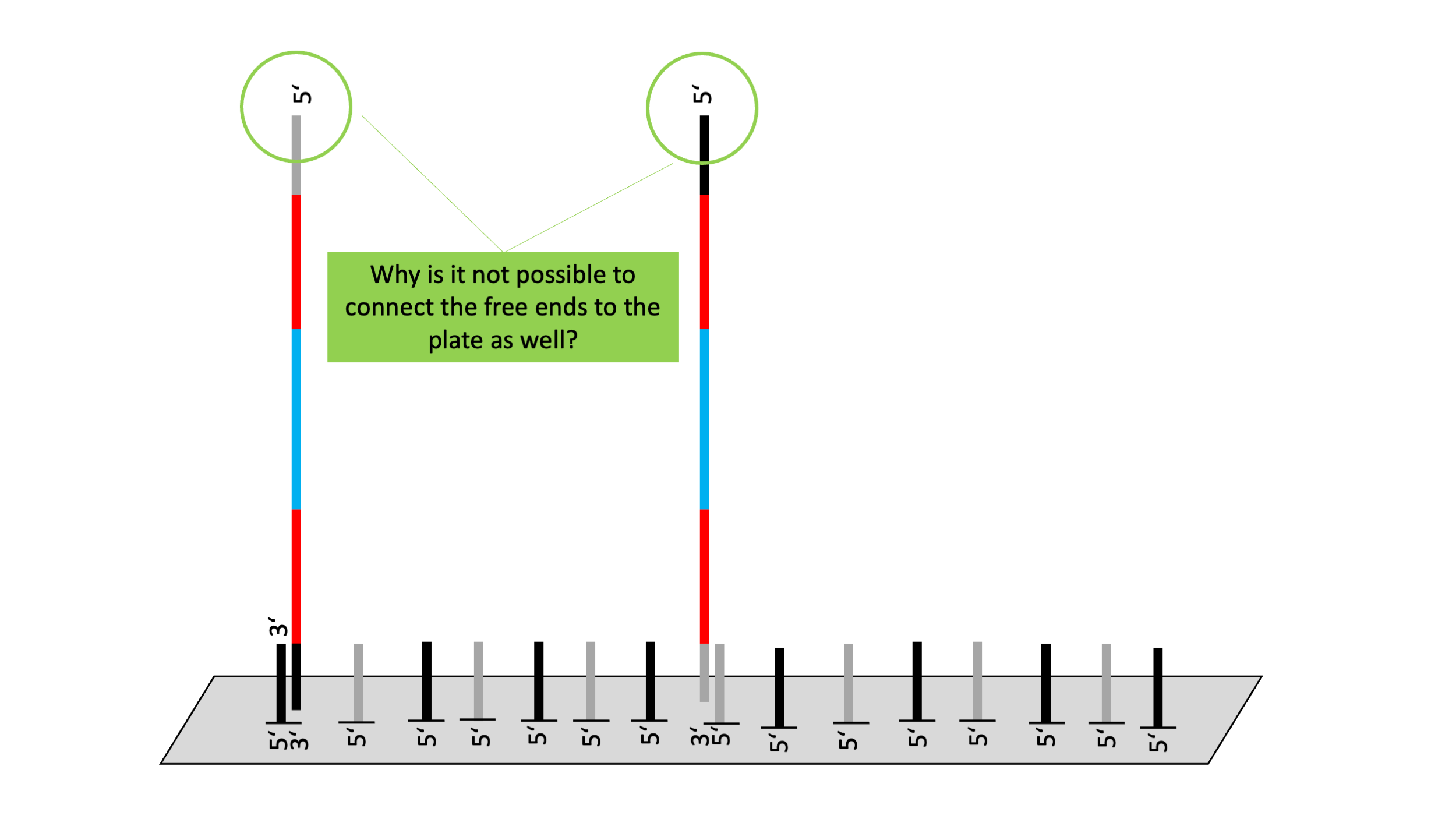
Binding of sequences to the flow cell
- Flow cell is a slide on which both capture sequences are attached.
- The attached capture sequences complement the adapters that have been ligated to the insert.





















The entire workflow as a video:
Take-home message
- Illumina uses sequencing by synthesis.
- Billions of DNA fragments (reads) are sequenced in parallel on a flow cell
- Large number of reads! But reads are short: 50 to 300 nucleotides
- Clusters are created through bridge PCR amplification
- Bases are detected by fluorescent labeled nucleotides
- Strength: high accuracy
- Limitation: short read length
