DNA and Sanger sequencing
Does this look familiar?

Desoxyribonucleic acid (DNA)
5’-ATGCACAACTTTTACAACTACAGCCCCCGCCCTGGGGCAAAACCTACGTGACGACAAT-3’ ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3’-TACGTGTTGAAAATGTTGATGTCGGGGGCGGGACCCCGTTTTGGATGCACTGCTGTTA-5’
- Biopolymer that codes for genetic information
- Monomers = nucleotides
- Bond types are phosphodiester bonds (3’-OH group)
- Complementary base pairing: (A=T; C≡G)
- Two antiparallel strands (running in opposite directions)
DNA replication fork

Direction of DNA
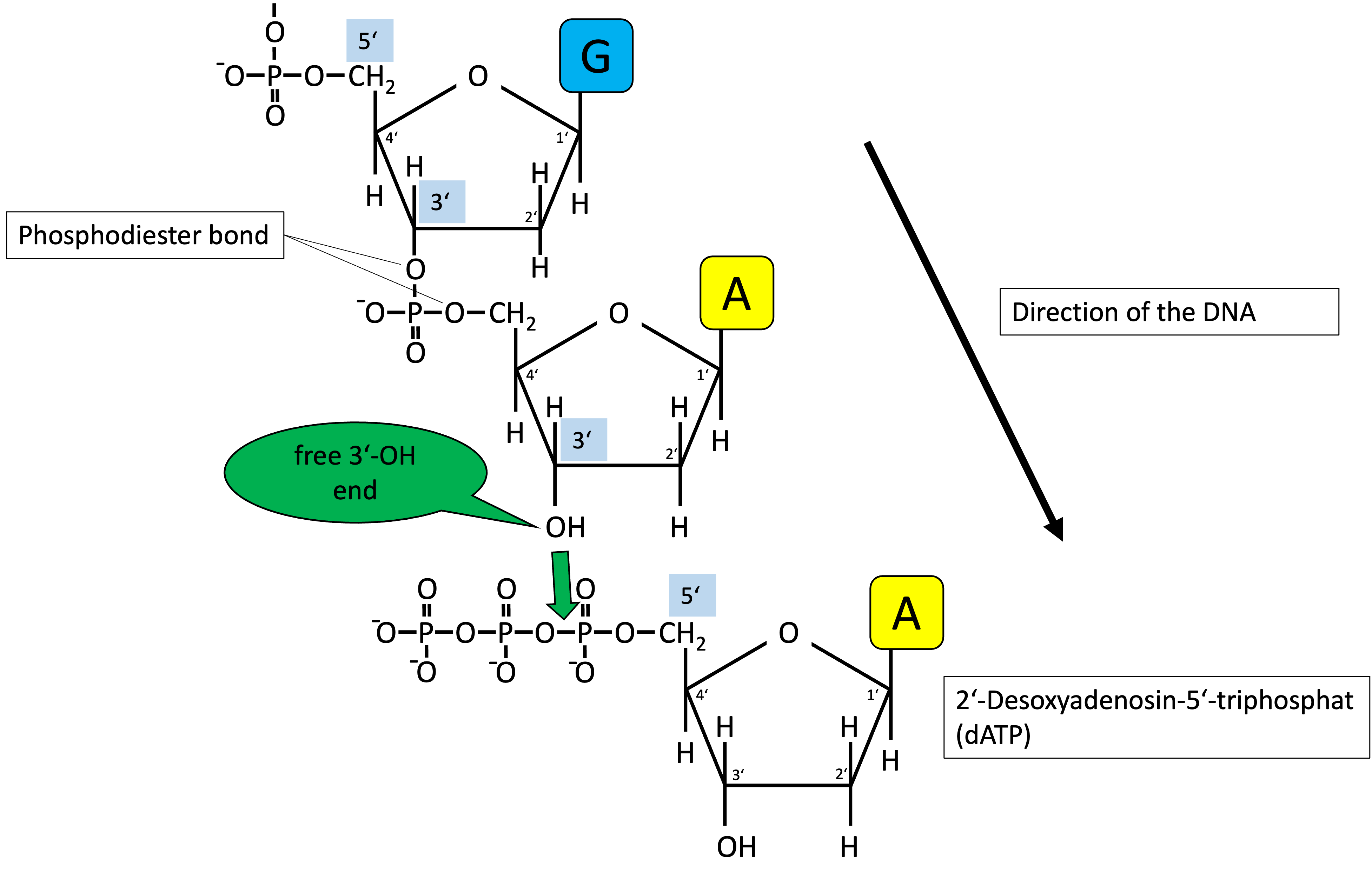
Four nucleotides
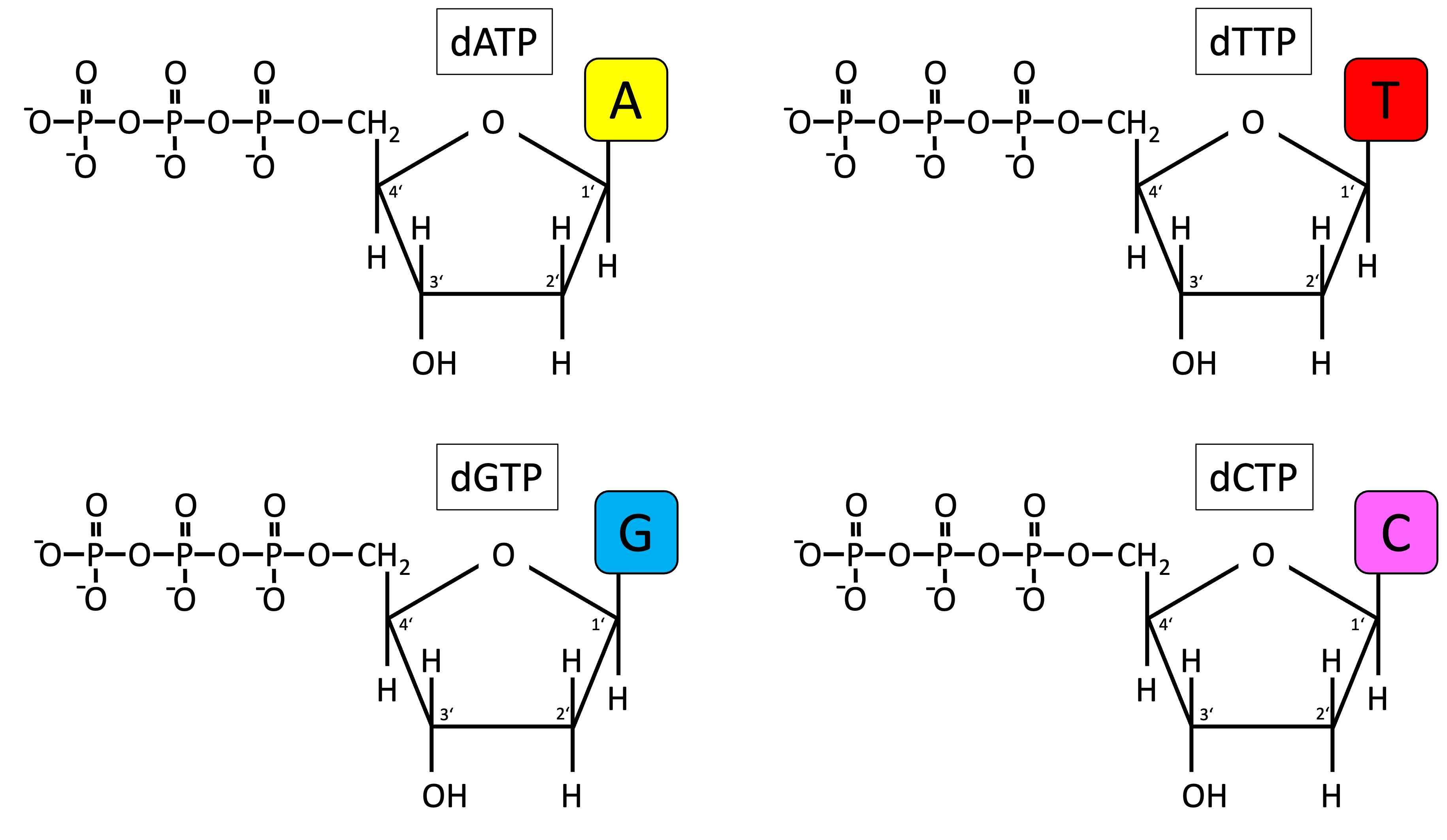
2’deoxyribose nucleoside triphosphate (dNTP)
2’,3’dideoxyribose nucleoside triphosphate (ddNTP)


Sanger sequencing
- based on modified nuclotides
- 2’,3’dideoxyribosee nucleoside triphoshate
- without 3’-OH group
- chain termination method


Sanger sequencing

Sanger sequencing chromatogram

Take-home message
Advantage of Sanger sequencing
- Devices are easy to use
- Results are obtained within couple of hours (overnight)
- Relatively cheep
Disadvantage of Sanger sequencing
- Few sequences are generated = low throughput technology
- Read length is limited to about 1000 nucleotides
